4m Read
3. joulukuuta 2024
RESPIRATORY HEALTH
Article
Proactive Surveillance to Ensure Broad Flu Strain Detection
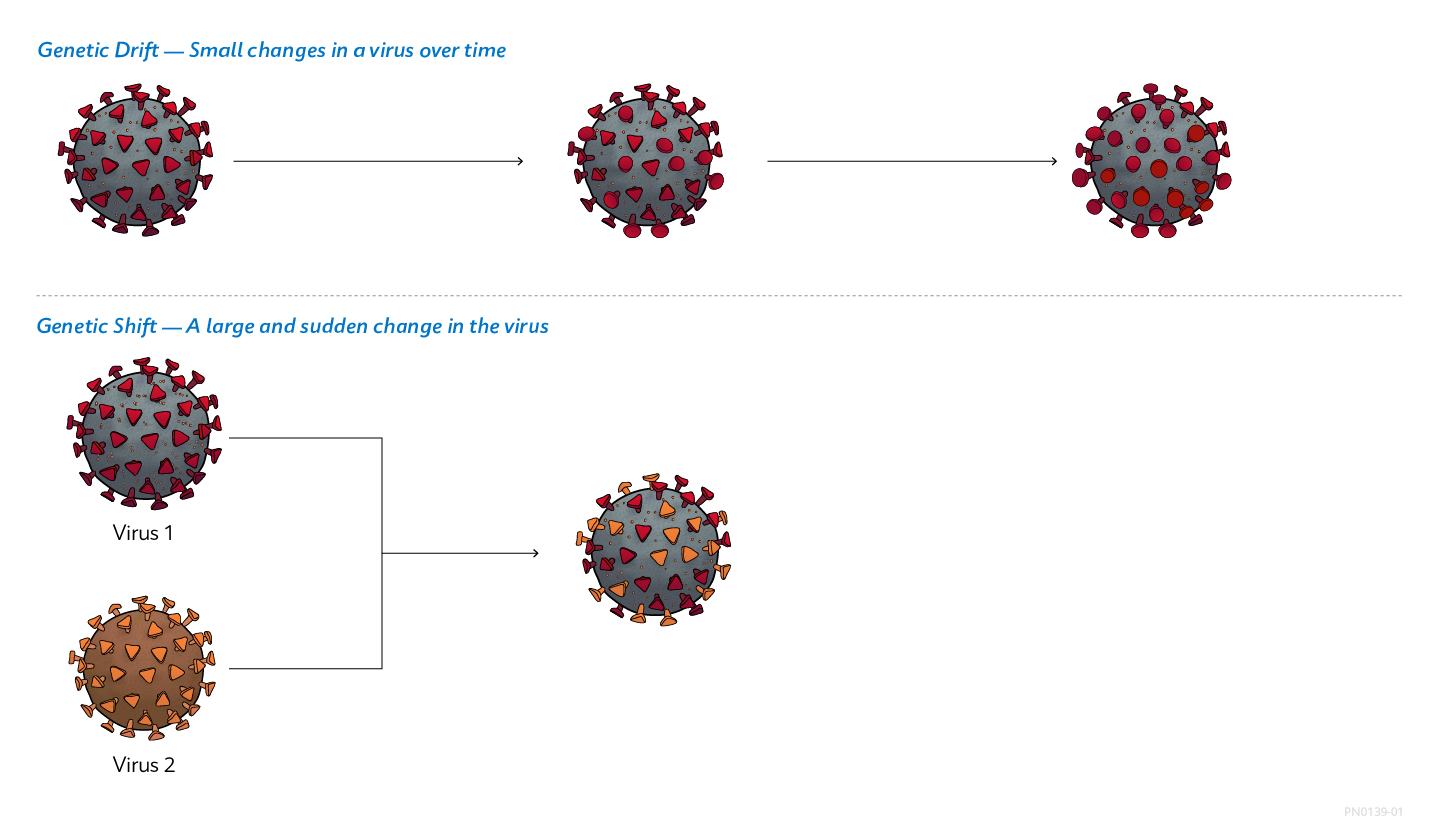
Viruses: Shifting and Drifting
Viruses constantly change by mutation. Two main mechanisms cause these changes. Genetic drift is a gradual change in a virus’ genetic makeup due to a series of minor genetic mutations.1 In influenza viruses, genetic drift results in small changes in viral surface proteins. Genetic shift, on the other hand, is an abrupt, major change in a virus’ genetic makeup that often creates a new strain. Genetic shift creates substantially different surface proteins and can give influenza viruses of animal origin the ability to infect humans, potentially causing outbreaks in human populations.1
Changes in viral genetic sequences can impact the performance of molecular diagnostic tests. If a test is designed to detect a particular viral sequence and that sequence changes due to mutation, the test could fail to detect the virus. Influenza, or flu virus, is prone to such mutations.
Genetic Drift and Shift
To avoid impacts on its PCR-based flu tests, Cepheid designs each test to detect multiple influenza genes. This reduces the impact of a mutation in a given genetic sequence as there are other unique sequences to enable detection (i.e., target redundancy). For influenza A, three gene targets are included to detect human-adapted strains and many avian strains. Additionally, two gene targets are included for influenza B.
Proactive Surveillance of Test Performance
Cepheid closely monitors strain surveillance data to stay updated on the evolving nature of influenza viruses. Cepheid uses in silico analysis as well as in vitro testing to monitor influenza strain coverage of its diagnostic tests. In silico analysis (i.e., experiments performed by computer) queries available sequence databases to predict the ability of a test to detect relevant influenza strains by assessing how well the PCR test sequences match the viral target sequences. Cepheid’s in silico analysis uses all available influenza sequences in the Global Initiative on Sharing All Influenza Data (GISAID) database (https://gisaid.org/). As of December 2023, over 18 000 human influenza A and over 7 000 human influenza B sequences were available and included in the analysis.
In vitro testing (experiments performed in a test tube or laboratory dish) of influenza virus isolates is also performed, using the annual Food and Drug Administration Center for Biologics Evaluation and Research (FDA CBER) Influenza Analytical Reactivity panel. The FDA CBER panel contains current human viruses including strains recommended by the World Health Organization for the annual flu vaccine.
To ensure continued influenza strain coverage and performance, Cepheid conducts in silico and in vitro analyses for its Xpert® Xpress Flu/RSV and Xpert Xpress CoV-2/Flu/RSV plus tests (collectively referred to here as Xpert Xpress). These multiplex in vitro diagnostic tests both contain multiple influenza targets.

Avian influenza A Strain Detection
With growing concerns about avian influenza “bird flu,” including recent human cases,2 avian strain coverage, including the H5N1 strain, is crucial for diagnostic tests. As of August 2023, over 5 000 avian influenza A subtype sequences were available in GISAID and included in Cepheid’s in silico analysis. Recent avian strains of concern, 2024 H5N1 Guangdong clade 2.3.4.4b, H5N1/A/Cambodia/NPH230032/2023 and H3N8/A/Guangdong/ZS-2023SF005/2023 were included.
Results from the combination of in silico analysis and in vitro testing demonstrate that the Xpert Xpress influenza tests will detect a wide range of current human and avian influenza virus strains, and is predicted to detect the highly pathogenic 2024 H5N1 Guangdong clade 2.3.4.4b that has been reported in outbreaks in dairy cattle and humans in the U.S.2-4
Outbreak Preparedness
The ability of a diagnostic test to accurately detect different strains of a virus can enable early detection and surveillance of novel or emerging strains, confident diagnoses for treatment and isolation measures, and potentially inform public health strategies for outbreak preparedness.
Continued data sharing, including the invaluable contributions of the scientific community that generated and shared influenza sequencing data via the GISAID Initiative, combined with ongoing proactive surveillance, will position public and global health initiatives to detect emerging strains early and avoid future outbreaks.
Contact us to request Cepheid’s Medical/Scientific Affairs Bulletin: Influenza Inclusivity of Cepheid Tests—2023 for a full report of the methods and results of this analysis.
Learn more about Cepheid's influenza tests here.
IVD. In vitro -diagnostinen lääkinnällinen laite. Ei välttämättä saatavilla kaikissa maissa.
Viitteet:
1. How Flu Viruses Can Change: “Drift” and “Shift” | CDC [Internet]. [cited 2024 Jul 10]. Available from: https://www.cdc.gov/flu/php/viruses/change.html?CDC_AAref_Val=https://www.cdc.gov/flu/about/viruses/change.htm
2. CDC Reports Fourth Human Case of H5 Bird Flu Tied to Dairy Cow Outbreak | CDC Online Newsroom | CDC [Internet]. [cited 2024 Jul 10]. Available from: https://www.cdc.gov/media/releases/2024/p-0703-4th-human-case-h5.html
3. Technical Report: June 2024 Highly Pathogenic Avian Influenza A(H5N1) Viruses | Bird Flu | CDC [Internet]. [cited 2024 Jul 10]. Available from: https://www.cdc.gov/bird-flu/php/technical-report/h5n1-06052024.html
4. Burrough ER, Magstadt DR, Petersen B, Timmermans SJ, Gauger PC, Zhang J, et al. Highly pathogenic avian influenza A(H5N1) clade 2.3.4.4b virus infection in domestic dairy cattle and cats, united states, 2024. Emerging Infect Dis. 2024 Jul;30(7):1335–43.
Read Next
LISÄÄ